ICESat-2 Altimeter Data using R
12 Feb 2022This blog post (which is a slight modification of both package Vignettes) explains the functionality of the IceSat2R R package and shows how to use the OpenAltimetry API from within R. It consists of three parts,
- OpenAltimetry
- IceSat-2 Mission Orbits
- IceSat-2 Atlas Products
OpenAltimetry
OpenAltimetry is a cyberinfrastructure platform for discovery, access, and visualization of data from NASA’s ICESat and ICESat-2 missions. These laser profiling altimeters are being used to measure changes in the topography of Earth’s,
- ice sheets
- vegetation canopy structure
- clouds and aerosols
The IceSat2R R package creates a connection to the OpenAltimetry API and allows the user (as of February 2022) to download and process the following ICESat-2 Altimeter Data,
- ‘ATL03’ (Global Geolocated Photon Data)
- ‘ATL06’ (Land Ice Height)
- ‘ATL07’ (Sea Ice Height)
- ‘ATL08’ (Land and Vegetation Height)
- ‘ATL10’ (Sea Ice Freeboard)
- ‘ATL12’ (Ocean Surface Height)
- ‘ATL13’ (Inland Water Surface Height)
The OpenAltimetry API restricts the requests to a 1x1 or 5x5 degree spatial bounding box, unless the “sampling” parameter is set to TRUE. The shiny application of the IceSat2R package allows the user to create a spatial grid of an AOI, preferably a 1- or 5-degree grid so that the selection can be within limits. An alternative would be to create a grid of smaller grid cells than required (for instance a 4-degree grid) and then to select multiple grid cells. The following .gif gives an idea of how this can be done from within Rstudio,
IceSat-2 Mission Orbits
This second part of the blog post is a slight modification of the first vignette and includes only the code snippets on how to download and process time specific orbits. The user can reproduce this second part by using the binder web-link of the IceSat2R R package or can go through the complete document (including the tables and visualizations) of the first vignette.
We’ll download one of the Reference Ground Track (RGT) cycles and merge it with other data sources with the purpose to visualize specific areas. We choose one of the latest which is “RGT_cycle_14” (from December 22, 2021 to March 23, 2022) and utilize 8 threads to speed up the pre-processing of the downloaded .kml files (the function takes approximately 15 minutes on my Linux Personal Computer),
require(IceSat2R)
require(magrittr)
require(sf)
sf::sf_use_s2(use_s2 = FALSE) # disable 's2' in this vignette
mapview::mapviewOptions(leafletHeight = '600px',
leafletWidth = '700px') # applies to all leaflet maps
avail_cycles = available_RGTs(only_cycle_names = TRUE)
avail_cycles
# [1] "RGT_cycle_1" "RGT_cycle_2" "RGT_cycle_3" "RGT_cycle_4" "RGT_cycle_5" "RGT_cycle_6"
# "RGT_cycle_7" "RGT_cycle_8" "RGT_cycle_9" "RGT_cycle_10" "RGT_cycle_11" "RGT_cycle_12"
# "RGT_cycle_13" "RGT_cycle_14" "RGT_cycle_15"
choose_cycle = avail_cycles[14]
choose_cycle
# [1] "RGT_cycle_14"
res_rgt_many = time_specific_orbits(RGT_cycle = choose_cycle,
download_method = 'curl',
threads = 8,
verbose = TRUE)
We’ll then create a data.table based on the coordinates, Date, Day of Year, Time and RGT,
rgt_subs = sf::st_coordinates(res_rgt_many)
colnames(rgt_subs) = c('longitude', 'latitude')
rgt_subs = data.table::data.table(rgt_subs)
rgt_subs$day_of_year = res_rgt_many$day_of_year
rgt_subs$Date = as.Date(res_rgt_many$Date_time)
rgt_subs$hour = lubridate::hour(res_rgt_many$Date_time)
rgt_subs$minute = lubridate::minute(res_rgt_many$Date_time)
rgt_subs$second = lubridate::second(res_rgt_many$Date_time)
rgt_subs$RGT = res_rgt_many$RGT
res_rgt_many = sf::st_as_sf(x = rgt_subs, coords = c('longitude', 'latitude'), crs = 4326)
res_rgt_many
Icesat-2 and Countries intersection
We’ll proceed to merge the orbit geometry points with the countries data of the rnaturalearth R package (1:110 million scales) and for this purpose, we keep only the “sovereignt” and “sov_a3” columns,
cntr = rnaturalearth::ne_countries(scale = 110, type = 'countries', returnclass = 'sf')
cntr = cntr[, c('sovereignt', 'sov_a3')]
cntr
We then merge the orbit points with the country geometries and specify also “left = TRUE” to keep also observations that do not intersect with the rnaturalearth countries data,
dat_both = suppressMessages(sf::st_join(x = res_rgt_many,
y = cntr,
join = sf::st_intersects,
left = TRUE))
dat_both
The unique number of RGT’s for “RGT_cycle_14” are
length(unique(dat_both$RGT))
We observe that from December 22, 2021 to March 23, 2022,
df_tbl = data.frame(table(dat_both$sovereignt), stringsAsFactors = F)
colnames(df_tbl) = c('country', 'Num_IceSat2_points')
df_subs = dat_both[, c('RGT', 'sovereignt')]
df_subs$geometry = NULL
df_subs = data.table::data.table(df_subs, stringsAsFactors = F)
colnames(df_subs) = c('RGT', 'country')
df_subs = split(df_subs, by = 'country')
df_subs = lapply(df_subs, function(x) {
unq_rgt = sort(unique(x$RGT))
items = ifelse(length(unq_rgt) < 5, length(unq_rgt), 5)
concat = paste(unq_rgt[1:items], collapse = '-')
iter_dat = data.table::setDT(list(country = unique(x$country),
Num_RGTs = length(unq_rgt),
first_5_RGTs = concat))
iter_dat
})
df_subs = data.table::rbindlist(df_subs)
df_tbl = merge(df_tbl, df_subs, by = 'country')
df_tbl = df_tbl[order(df_tbl$Num_IceSat2_points, decreasing = T), ]
DT_dtbl = DT::datatable(df_tbl, rownames = FALSE)
all RGT’s (1387 in number) intersect with “Antarctica” and almost all with “Russia”.
‘Onshore’ and ‘Offshore’ Points IceSat-2 coverage
The onshore and offshore number of IceSat-2 points and percentages for the “RGT_cycle_14” equal to
num_sea = sum(is.na(dat_both$sovereignt))
num_land = sum(!is.na(dat_both$sovereignt))
perc_sea = round(num_sea / nrow(dat_both), digits = 4) * 100.0
perc_land = round(num_land / nrow(dat_both), digits = 4) * 100.0
dtbl_land_sea = data.frame(list(percentage = c(perc_sea, perc_land),
Num_Icesat2_points = c(num_sea, num_land)))
row.names(dtbl_land_sea) = c('sea', 'land')
stargazer::stargazer(dtbl_land_sea,
type = 'html',
summary = FALSE,
rownames = TRUE,
header = FALSE,
table.placement = 'h',
title = 'Land and Sea Proportions')
Global glaciated areas and IceSat-2 coverage
We can also observe the IceSat-2 “RGT_cycle_14” coverage based on the 1 to 10 million large scale Natural Earth Glaciated Areas data,
ne_glaciers = system.file('data_files', 'ne_10m_glaciated_areas.RDS', package = "IceSat2R")
ne_obj = readRDS(file = ne_glaciers)
We’ll restrict the processing to the major polar glaciers (that have a name included),
ne_obj_subs = subset(ne_obj, !is.na(name))
ne_obj_subs = sf::st_make_valid(x = ne_obj_subs) # check validity of geometries
ne_obj_subs
and we’ll visualize the subset using the mapview package,
mpv = mapview::mapview(ne_obj_subs,
color = 'cyan',
col.regions = 'blue',
alpha.regions = 0.5,
legend = FALSE)
mpv
We will see which orbits of the IceSat-2 “RGT_cycle_14” intersect with these major polar glaciers,
res_rgt_many$id_rgt = 1:nrow(res_rgt_many) # include 'id' for fast subsetting
dat_glac_sf = suppressMessages(sf::st_join(x = ne_obj_subs,
y = res_rgt_many,
join = sf::st_intersects))
dat_glac = data.table::data.table(sf::st_drop_geometry(dat_glac_sf), stringsAsFactors = F)
dat_glac = dat_glac[complete.cases(dat_glac), ] # keep non-NA observations
dat_glac
We’ll split the merged data by the ‘name’ of the glacier,
dat_glac_name = split(x = dat_glac, by = 'name')
sum_stats_glac = lapply(dat_glac_name, function(x) {
dtbl_glac = x[, .(name_glacier = unique(name),
Num_unique_Dates = length(unique(Date)),
Num_unique_RGTs = length(unique(RGT)))]
dtbl_glac
})
sum_stats_glac = data.table::rbindlist(sum_stats_glac)
sum_stats_glac = sum_stats_glac[order(sum_stats_glac$Num_unique_RGTs, decreasing = T), ]
The next table shows the total number of days and RGTs for each one of the major polar glaciers,
stargazer::stargazer(sum_stats_glac,
type = 'html',
summary = FALSE,
rownames = FALSE,
header = FALSE,
table.placement = 'h',
title = 'Days and RGTs')
We can restrict to one of the glaciers to visualize the IceSat-2 “RGT_cycle_14” coverage over this specific area (‘Southern Patagonian Ice Field’),
sample_glacier = 'Southern Patagonian Ice Field'
dat_glac_smpl = dat_glac_name[[sample_glacier]]
cols_display = c('name', 'day_of_year', 'Date', 'hour', 'minute', 'second', 'RGT')
stargazer::stargazer(dat_glac_smpl[, ..cols_display],
type = 'html',
summary = FALSE,
rownames = FALSE,
header = FALSE,
table.placement = 'h',
title = 'Southern Patagonian Ice Field')
and we gather the intersected RGT coordinates points with the selected glacier,
subs_rgts = subset(res_rgt_many, id_rgt %in% dat_glac_smpl$id_rgt)
set.seed(1)
samp_colrs = sample(x = grDevices::colors(distinct = TRUE),
size = nrow(subs_rgts))
subs_rgts$color = samp_colrs
ne_obj_subs_smpl = subset(ne_obj_subs, name == sample_glacier)
mpv_glacier = mapview::mapview(ne_obj_subs_smpl,
color = 'cyan',
col.regions = 'blue',
alpha.regions = 0.5,
legend = FALSE)
mpv_RGTs = mapview::mapview(subs_rgts,
color = subs_rgts$color,
alpha.regions = 0.0,
lwd = 6,
legend = FALSE)
and visualize both the glacier and the subset of the intersected RGT coordinate points (of the different Days) in the same map. The clickable map and point popups include more information,
lft = mpv_glacier + mpv_RGTs
lft
IceSat-2 Atlas Products
This third part of the blog post relies on the computation of time specific orbits (as was the case in the first part) and explains how to use the ‘atl06’ ATLAS OpenAltimetry product. The complete third part (including the tables and interactive visualizations) can be found in HTML format in the mentioned web link. In the same way, as for the first part, the user can reproduce this second part by using the binder web-link of the IceSat2R R package.
We’ll continue using the Global glaciated areas and specifically, the Greenland Ice Sheet which is the second-largest ice body in the world, after the Antarctic ice sheet. The result of this third part will be a 3-dimensional visualization (you can have a look to the .gif at the end of the blog post).
Based on the OpenAltimetry documentation for the ATL06 (ATLAS/ICESat-2 L3A Land Ice Height, Version 4) Product, “This data set (ATL06) provides geolocated, land-ice surface heights (above the WGS 84 ellipsoid, ITRF2014 reference frame), plus ancillary parameters that can be used to interpret and assess the quality of the height estimates.”
We’ll use 2 different time periods of the year (winter and summer) to observe potential differences on the East part of the ‘Greenland Ice Sheet’ using the Land Ice Height (ATL06) Product and specifically we’ll use,
- for the winter (2020, 2021) the RGT_cycle_9 and RGT_cycle_10 (from 2020-12-15 to 2021-02-15)
- for the summer (2021) the RGT_cycle_11 and RGT_cycle_12 (from 2021-06-15 to 2021-08-15)
First, we’ll compute the time specific orbits for both periods,
require(IceSat2R)
require(magrittr)
require(sf)
sf::sf_use_s2(use_s2 = FALSE) # disable 's2' in this vignette
mapview::mapviewOptions(leafletHeight = '600px',
leafletWidth = '700px') # applies to all leaflet maps
#....................
# winter (2020, 2021)
#....................
start_date_w = "2020-12-15"
end_date_w = "2021-02-15"
rgt_winter = time_specific_orbits(date_from = start_date_w,
date_to = end_date_w,
RGT_cycle = NULL,
download_method = 'curl',
threads = parallel::detectCores(),
verbose = TRUE)
# ICESAT-2 orbits: 'Earliest-Date' is '2018-10-13' 'Latest-Date' is '2022-06-21'
# -----------------------------------------------------
# The .zip file of 'RGT_cycle_9' will be downloaded ...
# -----------------------------------------------------
# % Total % Received % Xferd Average Speed Time Time Time Current
# Dload Upload Total Spent Left Speed
# 100 140M 100 140M 0 0 3662k 0 0:00:39 0:00:39 --:--:-- 2904k
# The downloaded .zip file will be extracted in the '/tmp/RtmpPleeLI/RGT_cycle_9' directory ...
# Download and unzip the RGT_cycle_9 .zip file: Elapsed time: 0 hours and 0 minutes and 41 seconds.
# 138 .kml files will be processed ...
# Parallel processing of 138 .kml files using 8 threads starts ...
# The 'description' column of the output data will be processed ...
# The temproary files will be removed ...
# Processing of cycle 'RGT_cycle_9': Elapsed time: 0 hours and 1 minutes and 9 seconds.
# -----------------------------------------------------
# The .zip file of 'RGT_cycle_10' will be downloaded ...
# -----------------------------------------------------
# % Total % Received % Xferd Average Speed Time Time Time Current
# Dload Upload Total Spent Left Speed
# 100 140M 100 140M 0 0 3795k 0 0:00:37 0:00:37 --:--:-- 3841k
# The downloaded .zip file will be extracted in the '/tmp/RtmpPleeLI/RGT_cycle_10' directory ...
# Download and unzip the RGT_cycle_10 .zip file: Elapsed time: 0 hours and 0 minutes and 40 seconds.
# 824 .kml files will be processed ...
# Parallel processing of 824 .kml files using 8 threads starts ...
# The 'description' column of the output data will be processed ...
# The temproary files will be removed ...
# Processing of cycle 'RGT_cycle_10': Elapsed time: 0 hours and 7 minutes and 53 seconds.
# Total Elapsed time: 0 hours and 10 minutes and 25 seconds.
rgt_winter
# Simple feature collection with 91390 features and 14 fields
# Geometry type: POINT
# Dimension: XY
# Bounding box: xmin: -179.9029 ymin: -87.66478 xmax: 179.9718 ymax: 87.32805
# CRS: 4326
# First 10 features:
# Name timestamp begin end altitudeMode tessellate extrude visibility drawOrder icon RGT Date_time day_of_year cycle geometry
# 1 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:26:56 350 9 POINT (19.30252 0.01395099)
# 2 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:27:56 350 9 POINT (18.91822 3.854786)
# 3 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:28:56 350 9 POINT (18.53269 7.69588)
# 4 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:29:56 350 9 POINT (18.14468 11.53677)
# 5 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:30:56 350 9 POINT (17.75288 15.377)
# 6 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:31:56 350 9 POINT (17.3558 19.21615)
# 7 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:32:56 350 9 POINT (16.95175 23.05379)
# 8 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:33:56 350 9 POINT (16.53875 26.88954)
# 9 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:34:56 350 9 POINT (16.1144 30.72307)
# 10 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1250 2020-12-15 00:35:56 350 9 POINT (15.67575 34.55405)
for the winter period, it took approximately 10 min. to download and process the 962 .kml files utilizing 8 threads and then return an ‘sf’ (simple features) object. We’ll do the same for the summer period,
#..............
# summer (2021)
#..............
start_date_s = "2021-06-15"
end_date_s = "2021-08-15"
rgt_summer = time_specific_orbits(date_from = start_date_s,
date_to = end_date_s,
RGT_cycle = NULL,
download_method = 'curl',
threads = parallel::detectCores(),
verbose = TRUE)
# ICESAT-2 orbits: 'Earliest-Date' is '2018-10-13' 'Latest-Date' is '2022-06-21'
# -----------------------------------------------------
# The .zip file of 'RGT_cycle_11' will be downloaded ...
# -----------------------------------------------------
# % Total % Received % Xferd Average Speed Time Time Time Current
# Dload Upload Total Spent Left Speed
# 100 140M 100 140M 0 0 3440k 0 0:00:41 0:00:41 --:--:-- 4227k
# The downloaded .zip file will be extracted in the '/tmp/RtmpPleeLI/RGT_cycle_11' directory ...
# Download and unzip the RGT_cycle_11 .zip file: Elapsed time: 0 hours and 0 minutes and 44 seconds.
# 142 .kml files will be processed ...
# Parallel processing of 142 .kml files using 8 threads starts ...
# The 'description' column of the output data will be processed ...
# The temproary files will be removed ...
# Processing of cycle 'RGT_cycle_11': Elapsed time: 0 hours and 1 minutes and 30 seconds.
# -----------------------------------------------------
# The .zip file of 'RGT_cycle_12' will be downloaded ...
# -----------------------------------------------------
# % Total % Received % Xferd Average Speed Time Time Time Current
# Dload Upload Total Spent Left Speed
# 100 140M 100 140M 0 0 3204k 0 0:00:44 0:00:44 --:--:-- 2333k
# The downloaded .zip file will be extracted in the '/tmp/RtmpPleeLI/RGT_cycle_12' directory ...
# Download and unzip the RGT_cycle_12 .zip file: Elapsed time: 0 hours and 0 minutes and 47 seconds.
# 820 .kml files will be processed ...
# Parallel processing of 820 .kml files using 8 threads starts ...
# The 'description' column of the output data will be processed ...
# The temproary files will be removed ...
# Processing of cycle 'RGT_cycle_12': Elapsed time: 0 hours and 8 minutes and 20 seconds.
# Total Elapsed time: 0 hours and 11 minutes and 22 seconds.
rgt_summer
# Simple feature collection with 89965 features and 14 fields
# Geometry type: POINT
# Dimension: XY
# Bounding box: xmin: -179.9861 ymin: -87.66478 xmax: 179.9393 ymax: 87.32805
# CRS: 4326
# First 10 features:
# Name timestamp begin end altitudeMode tessellate extrude visibility drawOrder icon RGT Date_time day_of_year cycle geometry
# 1 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:12:24 166 11 POINT (-122.3822 0.06216933)
# 2 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:13:24 166 11 POINT (-122.7663 3.903004)
# 3 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:14:24 166 11 POINT (-123.1517 7.744086)
# 4 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:15:24 166 11 POINT (-123.5395 11.58495)
# 5 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:16:24 166 11 POINT (-123.9312 15.42515)
# 6 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:17:24 166 11 POINT (-124.3282 19.26424)
# 7 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:18:24 166 11 POINT (-124.7321 23.10183)
# 8 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:19:24 166 11 POINT (-125.145 26.93752)
# 9 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:20:24 166 11 POINT (-125.5693 30.77098)
# 10 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1256 2021-06-15 01:21:24 166 11 POINT (-126.0079 34.60189)
for the summer it took approximately the same time as for winter to process the 962 .kml files of the 2 Reference Ground Track (RGT) cycles. We’ll proceed to find the intersection of these 2 time periods (winter, summer) with the area of the East ‘Greenland Ice Sheet’,
#.......................................
# extract the 'Greenland Ice Sheet'
# glacier from the 'Natural Earth' data
#.......................................
ne_glaciers = system.file('data_files', 'ne_10m_glaciated_areas.RDS', package = "IceSat2R")
ne_obj = readRDS(file = ne_glaciers)
greenl_sh = subset(ne_obj, name == "Greenland Ice Sheet")
greenl_sh
We’ll continue with one of the 2 Greenland Ice Sheet parts (‘East’)
greenl_sh_east = greenl_sh[2, ]
# mapview::mapview(greenl_sh_east, legend = F)
#.....................................................
# create the bounding of the selected area because
# it's required for the 'OpenAltimetry' functions this
# will increase the size of the initial east area
#.....................................................
bbx_greenl_sh_east = sf::st_bbox(obj = greenl_sh_east)
sfc_bbx_greenl_sh_east = sf::st_as_sfc(bbx_greenl_sh_east)
# mapview::mapview(sfc_bbx_greenl_sh_east, legend = F)
#..............................................
# intersection with the computed "winter" RGT's
#..............................................
inters_winter = sf::st_intersects(x = sfc_bbx_greenl_sh_east,
y = sf::st_geometry(rgt_winter),
sparse = TRUE)
#.....................
# matched (RGT) tracks
#.....................
df_inters_winter = data.frame(inters_winter)
rgt_subs_winter = rgt_winter[df_inters_winter$col.id, , drop = FALSE]
rgt_subs_winter
# Simple feature collection with 1079 features and 14 fields
# Geometry type: POINT
# Dimension: XY
# Bounding box: xmin: -53.06014 ymin: 61.26295 xmax: -19.23143 ymax: 80.40264
# CRS: 4326
# First 10 features:
# Name timestamp begin end altitudeMode tessellate extrude visibility drawOrder icon RGT Date_time day_of_year cycle geometry
# 117 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1251 2020-12-15 02:22:14 350 9 POINT (-21.20408 80.22745)
# 207 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1252 2020-12-15 03:51:31 350 9 POINT (-35.55485 61.30102)
# 208 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1252 2020-12-15 03:52:31 350 9 POINT (-36.46073 65.10252)
# 209 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1252 2020-12-15 03:53:31 350 9 POINT (-37.58486 68.89803)
# 210 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1252 2020-12-15 03:54:31 350 9 POINT (-39.07057 72.68529)
# 211 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1252 2020-12-15 03:55:31 350 9 POINT (-41.21981 76.4593)
# 212 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1252 2020-12-15 03:56:31 350 9 POINT (-44.79278 80.20694)
# 977 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1260 2020-12-15 16:35:50 350 9 POINT (-31.5489 80.36163)
# 978 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1260 2020-12-15 16:36:50 350 9 POINT (-35.2132 76.61571)
# 979 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1260 2020-12-15 16:37:50 350 9 POINT (-37.4011 72.84247)
From the initial 91390 coordinate points, 1079 of the winter period intersect with the bounding box of our Area of Interest (AOI). We’ll do the same for the summer period,
#..............................................
# intersection with the computed "summer" RGT's
#..............................................
inters_summer = sf::st_intersects(x = sfc_bbx_greenl_sh_east,
y = sf::st_geometry(rgt_summer),
sparse = TRUE)
#.....................
# matched (RGT) tracks
#.....................
df_inters_summer = data.frame(inters_summer)
rgt_subs_summer = rgt_summer[df_inters_summer$col.id, , drop = FALSE]
rgt_subs_summer
# Simple feature collection with 1066 features and 14 fields
# Geometry type: POINT
# Dimension: XY
# Bounding box: xmin: -53.06014 ymin: 61.26295 xmax: -19.23143 ymax: 80.40264
# CRS: 4326
# First 10 features:
# Name timestamp begin end altitudeMode tessellate extrude visibility drawOrder icon RGT Date_time day_of_year cycle geometry
# 407 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1260 2021-06-15 07:55:33 166 11 POINT (-31.5489 80.36163)
# 408 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1260 2021-06-15 07:56:33 166 11 POINT (-35.2132 76.61571)
# 409 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1260 2021-06-15 07:57:33 166 11 POINT (-37.4011 72.84247)
# 410 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1260 2021-06-15 07:58:33 166 11 POINT (-38.90659 69.05564)
# 411 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1260 2021-06-15 07:59:33 166 11 POINT (-40.04214 65.2604)
# 412 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1260 2021-06-15 08:00:33 166 11 POINT (-40.95516 61.45908)
# 1062 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1267 2021-06-15 18:45:35 166 11 POINT (-29.84843 61.31864)
# 1063 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1267 2021-06-15 18:46:35 166 11 POINT (-30.75514 65.1202)
# 1064 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1267 2021-06-15 18:47:35 166 11 POINT (-31.88062 68.91576)
# 1065 <NA> <NA> <NA> clampToGround -1 0 1 NA <NA> 1267 2021-06-15 18:48:35 166 11 POINT (-33.36863 72.70305)
for the summer period, the intersected points are 1066. Based on these 2 intersected objects of winter and summer we’ll also have to find the common RGT’s that we’ll use for comparison purposes,
#...............................................
# compute the unique RGT's for summer and winter
#...............................................
unq_rgt_winter = unique(rgt_subs_winter$RGT)
unq_rgt_summer = unique(rgt_subs_summer$RGT)
dif_rgt = setdiff(unique(rgt_subs_winter$RGT), unique(rgt_subs_summer$RGT))
cat(glue::glue("Number of RGT winter: {length(unq_rgt_winter)}"), '\n')
# Number of RGT winter: 230
cat(glue::glue("Number of RGT summer: {length(unq_rgt_summer)}"), '\n')
# Number of RGT summer: 227
cat(glue::glue("Difference in RGT: {length(dif_rgt)}"), '\n')
# Difference in RGT: 3
#......................
# find the intersection
#......................
inters_rgts = intersect(unq_rgt_winter, unq_rgt_summer)
#...................................................
# create the subset data RGT's for summer and winter
#...................................................
idx_w = which(rgt_subs_winter$RGT %in% inters_rgts)
subs_rgt_winter = rgt_subs_winter[idx_w, , drop = F]
idx_s = which(rgt_subs_summer$RGT %in% inters_rgts)
subs_rgt_summer = rgt_subs_summer[idx_s, , drop = F]
Then we have to verify that the intersected time specific orbit RGT’s match the OpenAltimetry Tracks for both winter and summer,
#...................................................
# function to find which of the time specific RGT's
# match the OpenAltimetry Track-ID's
#...................................................
verify_RGTs = function(rgt_subs, bbx_aoi, verbose = FALSE) {
tracks_dates_RGT = list()
for (item in 1:nrow(rgt_subs)) {
dat_item = rgt_subs[item, , drop = F]
Date = as.Date(dat_item$Date_time)
op_tra = getTracks(minx = as.numeric(bbx_aoi['xmin']),
miny = as.numeric(bbx_aoi['ymin']),
maxx = as.numeric(bbx_aoi['xmax']),
maxy = as.numeric(bbx_aoi['ymax']),
date = as.character(Date),
outputFormat = 'csv',
download_method = 'curl',
verbose = FALSE)
date_obj = dat_item$Date_time
tim_rgt = glue::glue("Date: {date_obj} Time specific RGT: '{dat_item$RGT}'")
if (nrow(op_tra) > 0) {
inters_trc = intersect(op_tra$track, dat_item$RGT)
if (length(inters_trc) > 0) {
iter_op_trac = paste(op_tra$track, collapse = ', ')
if (verbose) cat(glue::glue("Row: {item} {tim_rgt} OpenAltimetry: '{iter_op_trac}'"), '\n')
num_subl = glue::glue("{as.character(Date)}_{inters_trc}")
tracks_dates_RGT[[num_subl]] = data.table::setDT(list(date = as.character(Date),
RGT = inters_trc))
}
}
else {
if (verbose) cat(glue::glue("{tim_rgt} without an OpenAltimetry match!"), '\n')
}
}
tracks_dates_RGT = data.table::rbindlist(tracks_dates_RGT)
return(tracks_dates_RGT)
}
#.........................................................
# we keep the relevant columns and remove duplicated
# Dates and RGTs to iterate over each pair of observations
#.........................................................
#.......
# winter
#.......
subs_rgt_w_trc = subs_rgt_winter[, c('RGT', 'Date_time')]
subs_rgt_w_trc$Date_time = as.character(as.Date(subs_rgt_w_trc$Date_time))
dups_w = which(duplicated(sf::st_drop_geometry(subs_rgt_w_trc)))
subs_rgt_w_trc = subs_rgt_w_trc[-dups_w, ]
ver_trc_winter = verify_RGTs(rgt_subs = subs_rgt_w_trc,
bbx_aoi = bbx_greenl_sh_east,
verbose = TRUE)
colnames(ver_trc_winter) = c('date_winter', 'RGT')
#.......
# summer
#.......
subs_rgt_s_trc = subs_rgt_summer[, c('RGT', 'Date_time')]
subs_rgt_s_trc$Date_time = as.character(as.Date(subs_rgt_s_trc$Date_time))
dups_s = which(duplicated(sf::st_drop_geometry(subs_rgt_s_trc)))
subs_rgt_s_trc = subs_rgt_s_trc[-dups_s, ]
ver_trc_summer = verify_RGTs(rgt_subs = subs_rgt_s_trc,
bbx_aoi = bbx_greenl_sh_east,
verbose = TRUE)
colnames(ver_trc_summer) = c('date_summer', 'RGT')
#.............
# merge by RGT
#.............
rgts_ws = merge(ver_trc_winter, ver_trc_summer, by = 'RGT')
rgts_ws
# RGT date_summer date_winter
# 1: 2 2021-06-23 2020-12-24
# 2: 10 2021-06-24 2020-12-24
# 3: 11 2021-06-24 2020-12-24
# 4: 17 2021-06-24 2020-12-25
# 5: 18 2021-06-24 2020-12-25
# ---
# 214: 1366 2021-06-22 2020-12-22
# 215: 1367 2021-06-22 2020-12-22
# 216: 1373 2021-06-22 2020-12-23
# 217: 1374 2021-06-22 2020-12-23
# 218: 1382 2021-06-23 2020-12-23
Now that we have the available RGT’s for the winter and summer Dates we have to create the 5-degree grid of the Greenland Ice sheet bounding box, because the ‘atl06’ OpenAltimetry product allows queries up to 5x5 degrees. Rather than a 5-degree grid, we will create a 4.5-degree to avoid any ‘over limit’ OpenAltimetry API errors,
greenl_grid = degrees_to_global_grid(minx = as.numeric(bbx_greenl_sh_east['xmin']),
maxx = as.numeric(bbx_greenl_sh_east['xmax']),
maxy = as.numeric(bbx_greenl_sh_east['ymax']),
miny = as.numeric(bbx_greenl_sh_east['ymin']),
degrees = 4.5,
square_geoms = TRUE,
crs_value = 4326,
verbose = TRUE)
In the beginning, we created the bounding box of the East ‘Greenland Ice Sheet’ (the bbx_greenl_sh_east object), which automatically increased the dimensions of the Area of Interest (AOI). Now, that we have the up to 5x5 degree grid we can keep the grid cells that intersect with our initial area,
inters_init = sf::st_intersects(sf::st_geometry(greenl_sh_east), greenl_grid)
inters_init = data.frame(inters_init)
inters_init = inters_init$col.id
greenl_grid_subs = greenl_grid[inters_init, , drop = F]
# Simple feature collection with 27 features and 1 field
# Geometry type: POLYGON
# Dimension: XY
# Bounding box: xmin: -53.10888 ymin: 60.11961 xmax: -17.10888 ymax: 82.61961
# CRS: EPSG:4326
# First 10 features:
# geometry area
# 1 POLYGON ((-53.10888 60.1196... 116709.30 [km^2]
# 2 POLYGON ((-48.60888 60.1196... 116709.30 [km^2]
# 3 POLYGON ((-44.10888 60.1196... 116709.30 [km^2]
# 9 POLYGON ((-53.10888 64.6196... 98928.06 [km^2]
# 10 POLYGON ((-48.60888 64.6196... 98928.06 [km^2]
# 11 POLYGON ((-44.10888 64.6196... 98928.06 [km^2]
# 12 POLYGON ((-39.60888 64.6196... 98928.06 [km^2]
# 13 POLYGON ((-35.10888 64.6196... 98928.06 [km^2]
# 14 POLYGON ((-30.60888 64.6196... 98928.06 [km^2]
# 15 POLYGON ((-26.10888 64.6196... 98928.06 [km^2]
We also have to make sure that the winter and summer data intersect with the up to 5x5 degree grid,
#............
# winter join
#............
subs_join_w = sf::st_join(x = greenl_grid_subs,
y = subs_rgt_w_trc,
join = sf::st_intersects,
left = FALSE)
subs_join_w = sf::st_as_sfc(unique(sf::st_geometry(subs_join_w)), crs = 4326)
subs_join_w
# Geometry set for 9 features
# Geometry type: POLYGON
# Dimension: XY
# Bounding box: xmin: -53.10888 ymin: 60.11961 xmax: -17.10888 ymax: 82.61961
# CRS: EPSG:4326
# First 5 geometries:
# POLYGON ((-53.10888 60.11961, -48.60888 60.1196...
# POLYGON ((-48.60888 60.11961, -44.10888 60.1196...
# POLYGON ((-44.10888 60.11961, -39.60888 60.1196...
# POLYGON ((-26.10888 73.61961, -21.60888 73.6196...
# POLYGON ((-21.60888 73.61961, -17.10888 73.6196...
#............
# summer join
#............
subs_join_s = sf::st_join(x = greenl_grid_subs,
y = subs_rgt_s_trc,
join = sf::st_intersects,
left = FALSE)
subs_join_s = sf::st_as_sfc(unique(sf::st_geometry(subs_join_s)), crs = 4326)
subs_join_s
# Geometry set for 9 features
# Geometry type: POLYGON
# Dimension: XY
# Bounding box: xmin: -53.10888 ymin: 60.11961 xmax: -17.10888 ymax: 82.61961
# CRS: EPSG:4326
# First 5 geometries:
# POLYGON ((-53.10888 60.11961, -48.60888 60.1196...
# POLYGON ((-48.60888 60.11961, -44.10888 60.1196...
# POLYGON ((-44.10888 60.11961, -39.60888 60.1196...
# POLYGON ((-26.10888 73.61961, -21.60888 73.6196...
# POLYGON ((-21.60888 73.61961, -17.10888 73.6196...
Since the winter and summer intersected spatial data are identical,
identical(subs_join_w, subs_join_s)
# [1] TRUE
we’ll iterate only over one of the two ‘sfc’ objects.
We can also visualize the available areas after the sf-join between the winter and summer spatial data and the East ‘Greenland Ice Sheet’,
mapview::mapview(subs_join_s, legend = F)
We can proceed that way and download the available ‘atl06’ Land Ice Height data using the get_level3a_data() function that takes a time interval and a bounding box as input (the bounding box will consist of up to a 5x5 degree grid cells). To reduce the computation time in this vignette we’ll restrict the for-loop to the first 5 Greenland Grid Cells,
join_geoms = 1:5
subs_join_reduced = subs_join_s[join_geoms]
mapview::mapview(subs_join_reduced, legend = F)
and to the following RGTs,
#...............................................
# keep a subset of RGTs and Greenland Grid cells
#...............................................
RGTs = c(33, 41, 56, 94, 108, 284,
421, 422, 437, 460, 475,
521, 544, 658, 681, 787,
794, 1290, 1366, 1373)
#......................
# update the input data
#......................
rgts_ws_reduced = subset(rgts_ws, RGT %in% RGTs)
rgts_ws_RGT = rgts_ws_reduced$RGT
#..........................................................
# we'll loop over the RGT's for the Date start and Date end
#..........................................................
# length(unique(rgts_ws_reduced$RGT)) == nrow(rgts_ws_reduced) # check for duplicated RGT's
rgts_ws_reduced$date_summer = as.Date(rgts_ws_reduced$date_summer)
rgts_ws_reduced$date_winter = as.Date(rgts_ws_reduced$date_winter)
min_max_dates = apply(rgts_ws_reduced[, -1], 2, function(x) c(min(x), max(x)))
start_w = as.character(min_max_dates[1, 'date_winter'])
end_w = as.character(min_max_dates[2, 'date_winter'])
start_s = as.character(min_max_dates[1, 'date_summer'])
end_s = as.character(min_max_dates[2, 'date_summer'])
dat_out_w = dat_out_s = logs_out = list()
LEN = length(subs_join_reduced)
LEN_rgt = length(rgts_ws_RGT)
t_start = proc.time()
for (idx_grid in 1:LEN) {
geom_iter = subs_join_reduced[idx_grid]
bbx_iter = sf::st_bbox(obj = geom_iter, crs = 4326)
for (j in 1:LEN_rgt) {
message("Greenland Geom: ", idx_grid, "/", LEN, " RGT-index: ", j, "/", LEN_rgt, "\r", appendLF = FALSE)
utils::flush.console()
track_i = rgts_ws_RGT[j]
name_iter = glue::glue("geom_idx_{idx_grid}_RGT_{track_i}")
iter_dat_winter = get_level3a_data(minx = as.numeric(bbx_iter['xmin']),
miny = as.numeric(bbx_iter['ymin']),
maxx = as.numeric(bbx_iter['xmax']),
maxy = as.numeric(bbx_iter['ymax']),
startDate = start_w,
endDate = end_w,
trackId = track_i,
beamName = NULL, # return data of all 6 beams
product = 'atl06',
client = 'portal',
outputFormat = 'csv',
verbose = FALSE)
iter_dat_summer = get_level3a_data(minx = as.numeric(bbx_iter['xmin']),
miny = as.numeric(bbx_iter['ymin']),
maxx = as.numeric(bbx_iter['xmax']),
maxy = as.numeric(bbx_iter['ymax']),
startDate = start_s,
endDate = end_s,
trackId = track_i,
beamName = NULL, # return data of all 6 beams
product = 'atl06',
client = 'portal',
outputFormat = 'csv',
verbose = FALSE)
NROW_w = nrow(iter_dat_winter)
NROW_s = nrow(iter_dat_summer)
if (NROW_s > 0 & NROW_w > 0) {
iter_logs = list(RGT = track_i,
N_rows_winter = NROW_w,
N_rows_summer = NROW_s)
logs_out[[name_iter]] = data.table::setDT(iter_logs)
dat_out_w[[name_iter]] = iter_dat_winter
dat_out_s[[name_iter]] = iter_dat_summer
}
}
}
IceSat2R:::compute_elapsed_time(time_start = t_start)
# Elapsed time: 0 hours and 6 minutes and 15 seconds.
We then sort and observe the output LOGs,
logs_out_dtbl = data.table::rbindlist(logs_out)
logs_out_dtbl$index = names(dat_out_w)
logs_out_dtbl = logs_out_dtbl[order(logs_out_dtbl$N_rows_winter, decreasing = T), ]
stargazer::stargazer(logs_out_dtbl,
type = 'html',
summary = FALSE,
rownames = FALSE,
header = FALSE,
table.placement = 'h',
title = 'LOGs')
We’ll first process and visualize one of Greenland’s geometries and RGT,
#................................
# we pick one with approx. same
# rows for both summer and winter
#................................
# names(dat_out_w)
Greenland_Geom_index = 4
RGT = 1290
sublist_name = glue::glue("geom_idx_{Greenland_Geom_index}_RGT_{RGT}")
#...............
# winter sublist
#...............
w_subs = dat_out_w[[sublist_name]]
w_subs
# date segment_id longitude latitude h_li atl06_quality_summary track_id beam file_name
# 1: 2020-12-17 566420 -22.43278 78.11961 553.5237 0 1290 gt1l processed_ATL06_20201217154357_12900905_005_01.h5
# 2: 2020-12-17 566421 -22.43294 78.11943 553.5601 0 1290 gt1l processed_ATL06_20201217154357_12900905_005_01.h5
# 3: 2020-12-17 566422 -22.43310 78.11926 553.5809 0 1290 gt1l processed_ATL06_20201217154357_12900905_005_01.h5
# 4: 2020-12-17 566423 -22.43326 78.11908 553.5331 0 1290 gt1l processed_ATL06_20201217154357_12900905_005_01.h5
# 5: 2020-12-17 566424 -22.43341 78.11891 553.5400 0 1290 gt1l processed_ATL06_20201217154357_12900905_005_01.h5
# ---
# 152558: 2020-12-17 591901 -25.70488 73.62039 1640.8195 0 1290 gt3r processed_ATL06_20201217154357_12900905_005_01.h5
# 152559: 2020-12-17 591902 -25.70496 73.62021 1650.2201 0 1290 gt3r processed_ATL06_20201217154357_12900905_005_01.h5
# 152560: 2020-12-17 591903 -25.70505 73.62003 1657.0470 0 1290 gt3r processed_ATL06_20201217154357_12900905_005_01.h5
# 152561: 2020-12-17 591904 -25.70514 73.61985 1665.8877 0 1290 gt3r processed_ATL06_20201217154357_12900905_005_01.h5
# 152562: 2020-12-17 591905 -25.70523 73.61968 1675.1165 0 1290 gt3r processed_ATL06_20201217154357_12900905_005_01.h5
#...............
# summer sublist
#...............
s_subs = dat_out_s[[sublist_name]]
s_subs
# date segment_id longitude latitude h_li atl06_quality_summary track_id beam file_name
# 1: 2021-06-17 566420 -22.43270 78.11961 554.1851 0 1290 gt1l processed_ATL06_20210617070339_12901105_005_01.h5
# 2: 2021-06-17 566421 -22.43286 78.11943 554.2042 0 1290 gt1l processed_ATL06_20210617070339_12901105_005_01.h5
# 3: 2021-06-17 566422 -22.43302 78.11925 554.1635 0 1290 gt1l processed_ATL06_20210617070339_12901105_005_01.h5
# 4: 2021-06-17 566423 -22.43318 78.11908 554.1546 0 1290 gt1l processed_ATL06_20210617070339_12901105_005_01.h5
# 5: 2021-06-17 566424 -22.43334 78.11890 554.2347 0 1290 gt1l processed_ATL06_20210617070339_12901105_005_01.h5
# ---
# 152048: 2021-06-17 591901 -25.70511 73.62039 1638.9667 0 1290 gt3r processed_ATL06_20210617070339_12901105_005_01.h5
# 152049: 2021-06-17 591902 -25.70521 73.62022 1646.6494 0 1290 gt3r processed_ATL06_20210617070339_12901105_005_01.h5
# 152050: 2021-06-17 591903 -25.70530 73.62004 1654.2173 0 1290 gt3r processed_ATL06_20210617070339_12901105_005_01.h5
# 152051: 2021-06-17 591904 -25.70539 73.61986 1663.3259 0 1290 gt3r processed_ATL06_20210617070339_12901105_005_01.h5
# 152052: 2021-06-17 591905 -25.70548 73.61968 1673.2574 0 1290 gt3r processed_ATL06_20210617070339_12901105_005_01.h5
The OpenAltimetry Data Dictionary includes the definitions for the column names of the output data.tables (except for the beam column which appears in the ‘level3a’ product of the OpenAltimetry API website),
- segment_id: “Segment number, counting from the equator. Equal to the segment_id for the second of the two 20m ATL03 segments included in the 40m ATL06 segment”
- h_li: “Standard land-ice segment height determined by land ice algorithm, corrected for first-photon bias, representing the median-based height of the selected ‘signal photon events’ (PEs)”
- atl06_quality_summary: “The ATL06_quality_summary parameter indicates the best-quality subset of all ATL06 data. A 0.0 (zero) in this parameter implies that no data-quality tests have found a problem with the segment, a 1.0 (one) implies that some potential problem has been found. Users who select only segments with zero values for this flag can be relatively certain of obtaining high-quality data, but will likely miss a significant fraction of usable data, particularly in cloudy, rough, or low-surface-reflectance conditions.”
- beam: the 6 (six) beams ‘gt1l’, ‘gt1r’, ‘gt2l’, ‘gt2r’, ‘gt3l’ or ‘gt3r’
The Date of the selected sublist for winter is ‘2020-12-17’ whereas for summer is ‘2021-06-17’. We’ll
- exclude the low-quality observations using the ‘atl06_quality_summary’ column
- keep specific columns (‘date’, ‘segment_id’, ‘longitude’, ‘latitude’, ‘h_li’, ‘beam’)
- rename the winter and summer columns by adding the ‘_winter’ and ‘_summer’ extension
- merge the remaining observations (winter, summer) based on the ‘segment_id’ and ‘beam’ columns
- create an additional column (‘dif_height’) with the difference in height between the ‘h_li_winter’ and ‘h_li_summer’
cols_keep = c('date', 'segment_id', 'longitude', 'latitude', 'h_li', 'beam')
w_subs_hq = subset(w_subs, atl06_quality_summary == 0)
w_subs_hq = w_subs_hq[, ..cols_keep]
colnames(w_subs_hq) = glue::glue("{cols_keep}_winter")
s_subs_hq = subset(s_subs, atl06_quality_summary == 0)
s_subs_hq = s_subs_hq[, ..cols_keep]
colnames(s_subs_hq) = glue::glue("{cols_keep}_summer")
sw_hq_merg = merge(x = w_subs_hq,
y = s_subs_hq,
by.x = c('segment_id_winter', 'beam_winter'),
by.y = c('segment_id_summer', 'beam_summer'))
sw_hq_merg$dif_height = sw_hq_merg$h_li_winter - sw_hq_merg$h_li_summer
summary(sw_hq_merg$dif_height)
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# -42.6563 -0.4222 -0.0854 -0.1567 0.0537 26.4173
the following code snippet will create the visualizations for the beams “gt1l” and “gt1r” for the selected subset,
cols_viz = c('segment_id_winter', 'beam_winter', 'h_li_winter', 'h_li_summer')
ws_vis = sw_hq_merg[, ..cols_viz]
ws_vis_mlt = reshape2::melt(ws_vis, id.vars = c('segment_id_winter', 'beam_winter'))
ws_vis_mlt = data.table::data.table(ws_vis_mlt, stringsAsFactors = F)
ws_vis_mlt_spl = split(ws_vis_mlt, by = 'beam_winter')
# BEAMS = names(ws_vis_mlt_spl) # plot all beams
#...................................
# function to plot each subplot beam
#...................................
plotly_beams = function(spl_data,
beam,
left_width,
left_height,
right_width,
right_height) {
subs_iter = spl_data[[beam]]
cat(glue::glue("Plot for Beam '{beam}' will be created ..."), '\n')
#......................
# plot for all segments
#......................
fig_lines = plotly::plot_ly(data = subs_iter,
x = ~segment_id_winter,
y = ~value,
color = ~variable,
colors = c("blue", "red"),
line = list(width = 2),
text = ~glue::glue("land-ice-height: {value} Segment-id: {segment_id_winter}"),
hoverinfo = "text",
width = left_width,
height = left_height) %>%
plotly::layout(xaxis = list(gridcolor = "grey", showgrid = T),
yaxis = list(gridcolor = "grey", showgrid = T)) %>%
plotly::add_lines()
#..............................
# plot for a subset of segments
#..............................
segm_ids = 588326:588908 # this subset of segments show a big difference betw. summer and winter
subs_iter_segm = subset(subs_iter, segment_id_winter %in% segm_ids)
fig_lines_splt = plotly::plot_ly(data = subs_iter_segm,
x = ~segment_id_winter,
y = ~value,
color = ~variable,
colors = c("blue", "red"),
line = list(width = 2),
text = ~glue::glue("land-ice-height: {value} Segment-id: {segment_id_winter}"),
hoverinfo = "text",
width = right_width,
height = right_height) %>%
plotly::layout(xaxis = list(gridcolor = "grey", showgrid = T),
yaxis = list(gridcolor = "grey", showgrid = T)) %>%
plotly::add_lines(showlegend = FALSE)
both_plt = plotly::subplot(list(fig_lines, fig_lines_splt), nrows=1, margin = 0.03, widths = c(0.7, 0.3)) %>%
plotly::layout(title = glue::glue("Beam: '{beam}' ( Segments: from {min(segm_ids)} to {max(segm_ids)} )"))
# plotly::export(p = both_plt, file = glue::glue('{beam}.png'))
return(both_plt)
}
The output left plot shows all segment-id’s for the “gt1l” and “gt1r” beams, whereas the right plot is restricted only to the segment-id’s from 588326 to 588908 to highlight potential differences in land-ice-height between the winter and summer periods,
plt_gt1l = plotly_beams(spl_data = ws_vis_mlt_spl,
beam = "gt1l",
left_width = 1800,
left_height = 800,
right_width = 900,
right_height = 400)
plt_gt1l
plt_gt1r = plotly_beams(spl_data = ws_vis_mlt_spl,
beam = "gt1r",
left_width = 1800,
left_height = 800,
right_width = 900,
right_height = 400)
plt_gt1r
#......................................................
# save all images for all beams in a separate directory
#......................................................
nams_ws = names(dat_out_w)
save_summary = save_dat = list()
for (nam_iter in nams_ws) {
cat("-----------------\n")
cat(nam_iter, '\n')
cat("-----------------\n")
w_subs = dat_out_w[[nam_iter]]
s_subs = dat_out_s[[nam_iter]]
cols_keep = c('date', 'segment_id', 'longitude', 'latitude', 'h_li', 'beam')
w_subs_hq = subset(w_subs, atl06_quality_summary == 0)
w_subs_hq = w_subs_hq[, ..cols_keep]
colnames(w_subs_hq) = glue::glue("{cols_keep}_winter")
s_subs_hq = subset(s_subs, atl06_quality_summary == 0)
s_subs_hq = s_subs_hq[, ..cols_keep]
colnames(s_subs_hq) = glue::glue("{cols_keep}_summer")
sw_hq_merg = merge(x = w_subs_hq,
y = s_subs_hq,
by.x = c('segment_id_winter', 'beam_winter'),
by.y = c('segment_id_summer', 'beam_summer'))
if (nrow(sw_hq_merg) > 0) {
sw_hq_merg$dif_height = sw_hq_merg$h_li_winter - sw_hq_merg$h_li_summer
save_dat[[nam_iter]] = sw_hq_merg
save_summary[[nam_iter]] = data.table::setDT(list(name_iter = nam_iter,
min = min(sw_hq_merg$dif_height),
mean = mean(sw_hq_merg$dif_height),
median = median(sw_hq_merg$dif_height),
max = max(sw_hq_merg$dif_height),
N_rows = nrow(sw_hq_merg)))
#.......................................
# save the plots for visual verification
#.......................................
cols_viz = c('segment_id_winter', 'beam_winter', 'h_li_winter', 'h_li_summer')
ws_vis = sw_hq_merg[, ..cols_viz]
ws_vis_mlt = reshape2::melt(ws_vis, id.vars = c('segment_id_winter', 'beam_winter'))
ws_vis_mlt = data.table::data.table(ws_vis_mlt, stringsAsFactors = F)
ws_vis_mlt_spl = split(ws_vis_mlt, by = 'beam_winter')
dir_save = file.path('all_beams_all_RGTs', nam_iter) # !! create the 'all_beams_all_RGTs' directory first
if (!dir.exists(dir_save)) dir.create(dir_save)
BEAMS = names(ws_vis_mlt_spl) # plot all beams
for (beam in BEAMS) {
subs_iter = ws_vis_mlt_spl[[beam]]
cat(glue::glue("Plot for Beam '{beam}' will be saved ..."), '\n')
#......................
# plot for all segments
#......................
fig_lines = plotly::plot_ly(data = subs_iter,
x = ~segment_id_winter,
y = ~value,
color = ~variable,
colors = c("blue", "red"),
line = list(width = 2),
text = ~glue::glue("land-ice-height: {value} Segment-id: {segment_id_winter}"),
hoverinfo = "text",
width = 1800,
height = 1000) %>%
plotly::layout(xaxis = list(gridcolor = "grey", showgrid = T),
yaxis = list(gridcolor = "grey", showgrid = T)) %>%
plotly::add_lines()
plotly::export(p = fig_lines, file = file.path(dir_save, glue::glue('{beam}.png')))
}
}
else {
message(glue::glue("Empty data table after merging for idx and RGT: '{nam_iter}'"))
}
}
save_summary = data.table::rbindlist(save_summary)
save_summary = save_summary[order(save_summary$max, decreasing = T), ]
save_summary
Finally, we can also add the elevation of the AOI for comparison purposes. We’ll choose another Greenland Grid cell, RGT, and beams,
Greenland_Geom_index = 2
RGT = 33
sublist_name = glue::glue("geom_idx_{Greenland_Geom_index}_RGT_{RGT}")
w_subs = dat_out_w[[sublist_name]]
s_subs = dat_out_s[[sublist_name]]
cols_keep = c('date', 'segment_id', 'longitude', 'latitude', 'h_li', 'beam')
w_subs_hq = subset(w_subs, atl06_quality_summary == 0)
w_subs_hq = w_subs_hq[, ..cols_keep]
colnames(w_subs_hq) = glue::glue("{cols_keep}_winter")
s_subs_hq = subset(s_subs, atl06_quality_summary == 0)
s_subs_hq = s_subs_hq[, ..cols_keep]
colnames(s_subs_hq) = glue::glue("{cols_keep}_summer")
sw_hq_merg = merge(x = w_subs_hq,
y = s_subs_hq,
by.x = c('segment_id_winter', 'beam_winter'),
by.y = c('segment_id_summer', 'beam_summer'))
After merging the winter and summer data for the specific Grid Cell and RGT, I have to keep only one pair of (latitude, longitude) coordinates for visualization purposes. I’ll compute the distance between the winter and summer coordinates (of each row) for the same segment_id and beam and I’ll continue with the beams and observations that have the lowest difference in distance (for a fair comparison),
#...............................
# compute the pair-wise distance
#...............................
sw_hq_merg$dist_dif = geodist::geodist(x = sw_hq_merg[, c('longitude_winter', 'latitude_winter')],
y = sw_hq_merg[, c('longitude_summer', 'latitude_summer')],
paired = TRUE,
measure = 'geodesic')
#..............
# split by beam
#..............
spl_beam = split(sw_hq_merg, by = 'beam_winter')
#...............................................
# compute the summary of the distance to observe
# the beams with the lowest distance difference
#...............................................
sm_stats = lapply(spl_beam, function(x) {
summary(x$dist_dif)
})
# $gt1l
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 0.006829 1.568755 2.107793 2.083547 2.654482 3.911961
# $gt1r
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 0.0008427 0.2483260 0.5219864 0.5766648 0.8521821 2.0041663
# $gt2l
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 0.0002195 0.3001418 0.6112493 0.6753225 0.9860298 2.2545978
# $gt2r
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 3.170 4.697 5.356 5.300 5.931 7.170
# $gt3l
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 1.197 2.679 3.115 3.155 3.621 5.069
# $gt3r
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 3.881 5.066 5.488 5.498 5.949 7.114
We observe that the beams ‘gt1r’ and ‘gt2l’ return the lowest difference in distance (approximately 2.0 and 2.25 meters) for the coordinates between the ‘winter’ and ‘summer’ observations, therefore we continue with these 2 beams. Moreover, the ‘gt1r’ and ‘gt2l’ beams are separated by approximately 3 kilometers from each other.
#......................................
# keep only the 'gt1r' and 'gt2l' beams
#......................................
sw_hq_merg_beams = subset(sw_hq_merg, beam_winter %in% c('gt1r', 'gt2l'))
#.......................................................
# keep only a pair of coordinates and rename the columns
#.......................................................
sw_hq_merg_beams = sw_hq_merg_beams[, c('segment_id_winter', 'beam_winter', 'longitude_winter', 'latitude_winter', 'h_li_winter', 'h_li_summer')]
colnames(sw_hq_merg_beams) = c('segment_id', 'beam', 'longitude', 'latitude', 'h_li_winter', 'h_li_summer')
# segment_id beam longitude latitude h_li_winter h_li_summer
# 1: 351810 gt1r -44.10891 63.26390 2724.035 2725.023
# 2: 351811 gt1r -44.10895 63.26408 2724.004 2725.155
# 3: 351812 gt1r -44.10900 63.26426 2724.445 2725.298
# 4: 351813 gt1r -44.10904 63.26444 2724.530 2725.461
# 5: 351814 gt1r -44.10908 63.26462 2725.084 2725.650
# ---
# 8700: 359399 gt1r -44.44637 64.61873 2777.249 2777.390
# 8701: 359400 gt1r -44.44642 64.61891 2777.136 2777.297
# 8702: 359401 gt1r -44.44647 64.61908 2777.081 2777.192
# 8703: 359402 gt1r -44.44651 64.61926 2776.936 2777.076
# 8704: 359403 gt1r -44.44656 64.61944 2776.818 2776.968
Then we’ll download the 30-meter raster DEM (Digital Elevation Model) for the AOI using the CopernicusDEM R package,
sf_aoi = sf::st_as_sf(sw_hq_merg_beams, coords = c('longitude', 'latitude'), crs = 4326)
bbx_aoi = sf::st_bbox(sf_aoi)
sfc_aoi = sf::st_as_sfc(bbx_aoi)
dem_dir = tempdir()
dem_dir
dem30 = CopernicusDEM::aoi_geom_save_tif_matches(sf_or_file = sfc_aoi,
dir_save_tifs = dem_dir,
resolution = 30,
crs_value = 4326,
threads = parallel::detectCores(),
verbose = TRUE)
if (nrow(dem30$csv_aoi) > 1) { # create a .VRT file if I have more than 1 .tif files
file_out = file.path(dem_dir, 'VRT_mosaic_FILE.vrt')
vrt_dem30 = CopernicusDEM::create_VRT_from_dir(dir_tifs = dem_dir,
output_path_VRT = file_out,
verbose = TRUE)
}
if (nrow(dem30$csv_aoi) == 1) { # if I have a single .tif file keep the first index
file_out = list.files(dem_dir, pattern = '.tif', full.names = T)[1]
}
#.......................................................
# crop the raster to the bounding box of the coordinates
#.......................................................
rst_inp = terra::rast(x = file_out)
vec_crop = terra::vect(x = sfc_aoi)
rst_crop = terra::crop(x = rst_inp,
y = vec_crop,
snap = "out") # snap = "in" gives NA's
#...............................................
# we also have to find the closest elevation
# value to the 'winter' and 'summer' coordinates
# using the raster resolution
#...............................................
ter_dtbl = data.table::as.data.table(x = rst_crop, xy = TRUE, cells = TRUE)
colnames(ter_dtbl) = c("cell", "lon_dem30", "lat_dem30", "dem30")
# length(unique(ter_dtbl$cell)) == nrow(ter_dtbl)
xy = as.matrix(sw_hq_merg_beams[, c('longitude', 'latitude')])
sw_cells = terra::cellFromXY(object = rst_crop, xy = xy)
sw_hq_merg_beams$cell = sw_cells
# length(unique(sw_hq_merg_beams$cell)) < nrow(sw_hq_merg_beams)
merg_cells = merge(x = sw_hq_merg_beams, y = ter_dtbl, by = 'cell')
#......................................................................
# compute also the difference in distance between the beam measurements
# and the DEM coordinates (based on the 30-meter resolution cells)
#......................................................................
merg_cells$dem_dif_dist = geodist::geodist(x = merg_cells[, c('longitude', 'latitude')],
y = merg_cells[, c('lon_dem30', 'lat_dem30')],
paired = TRUE,
measure = 'geodesic')
summary(merg_cells$dem_dif_dist)
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 0.2356 8.1619 11.5943 11.1490 14.3201 20.5394
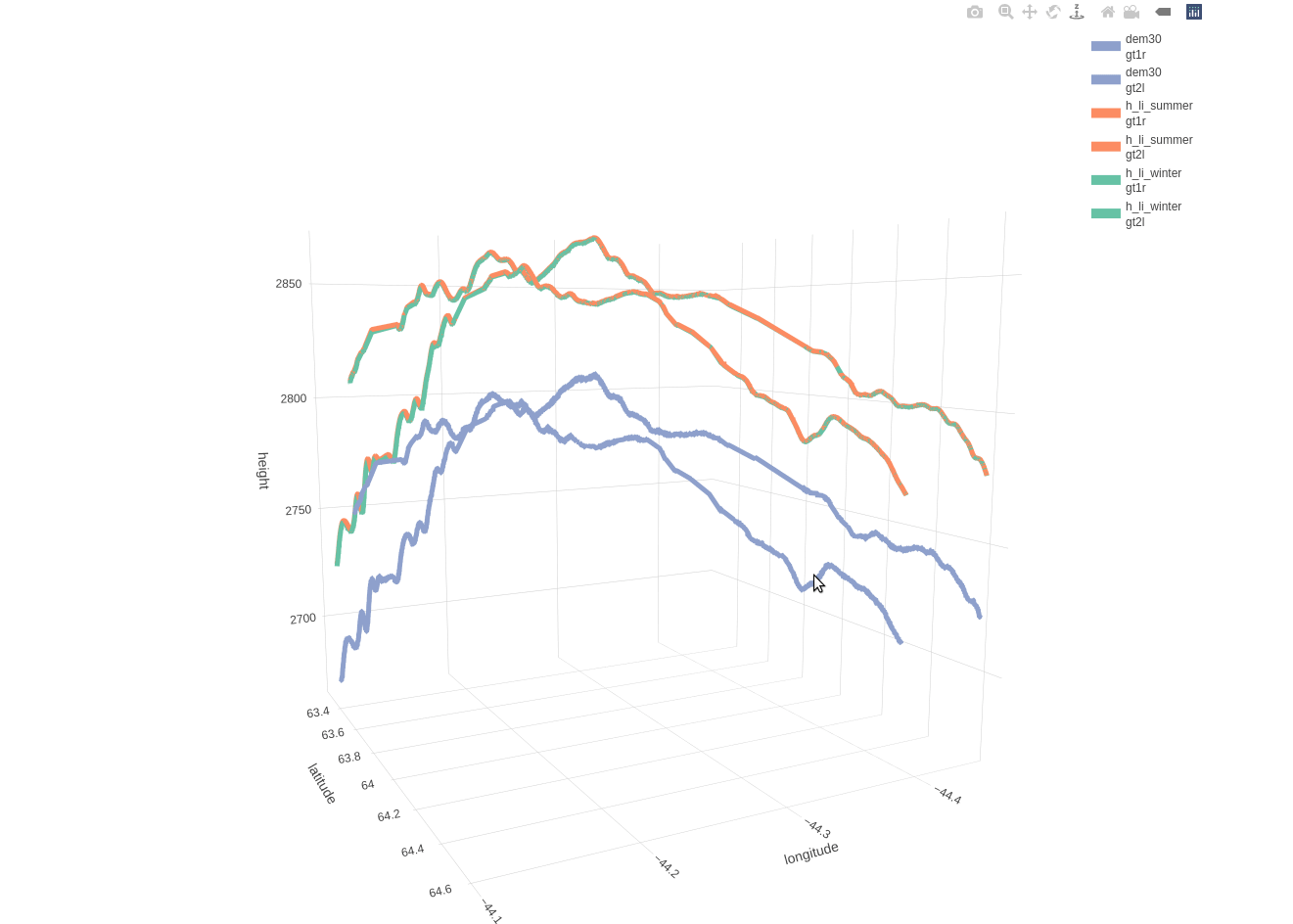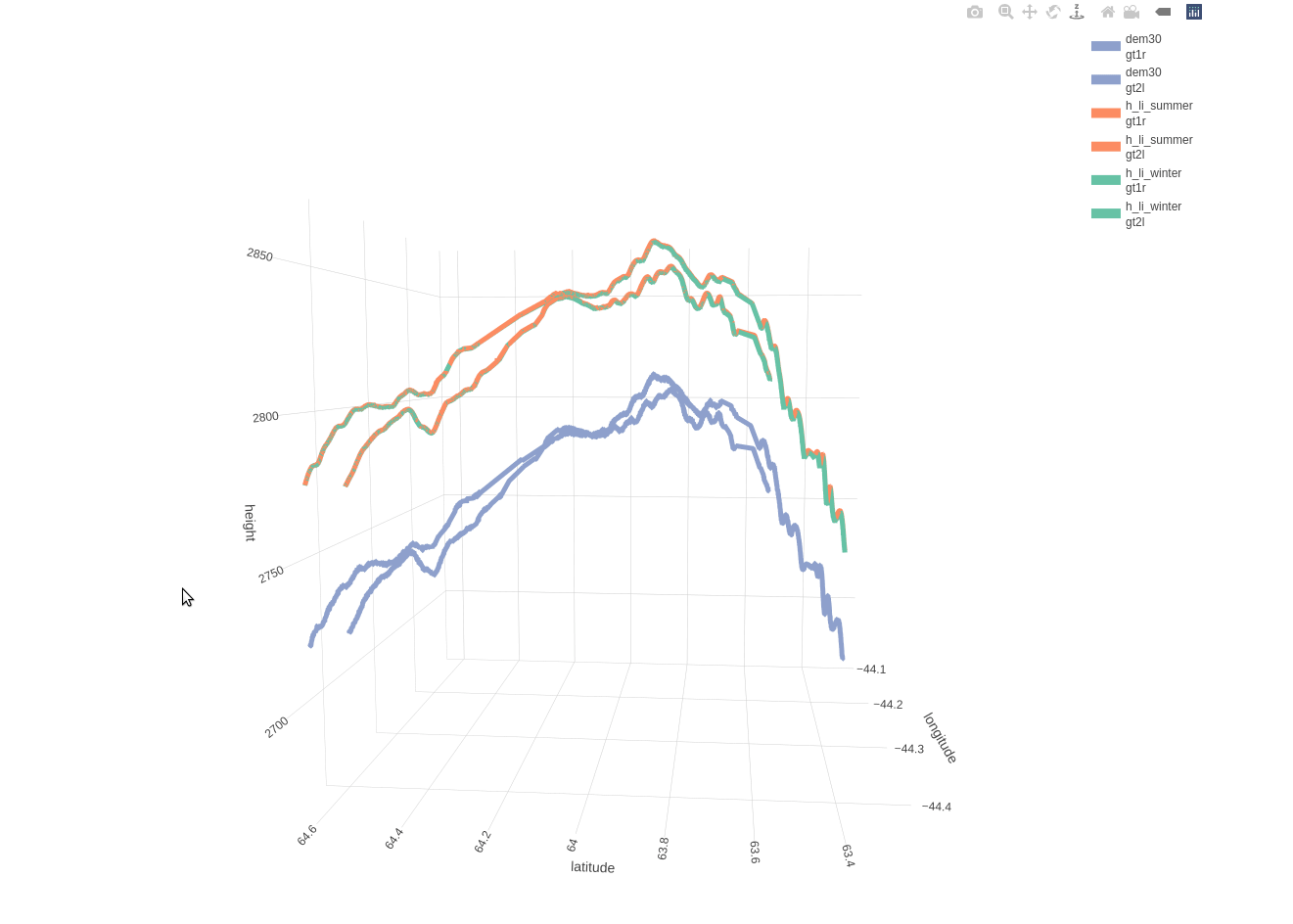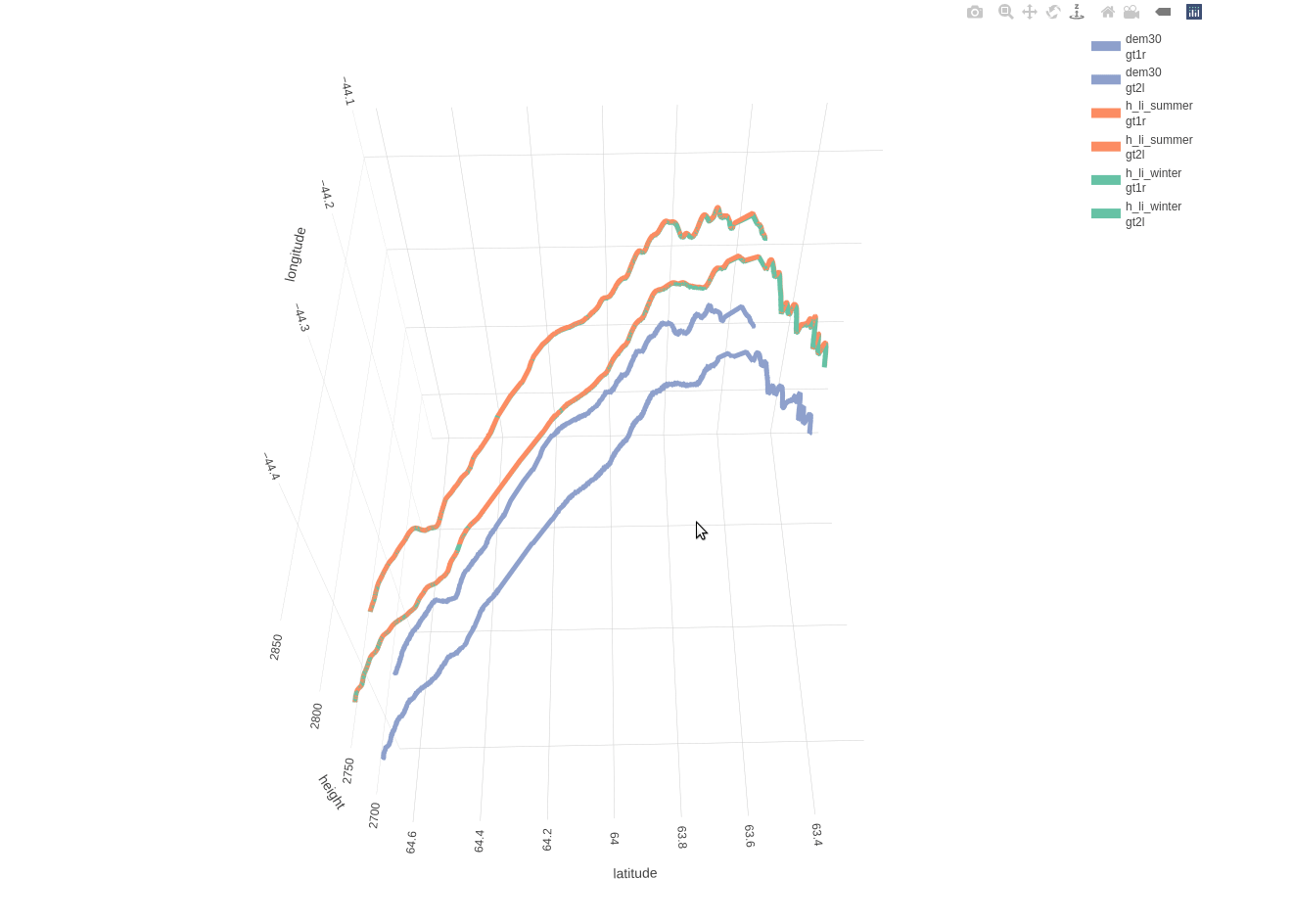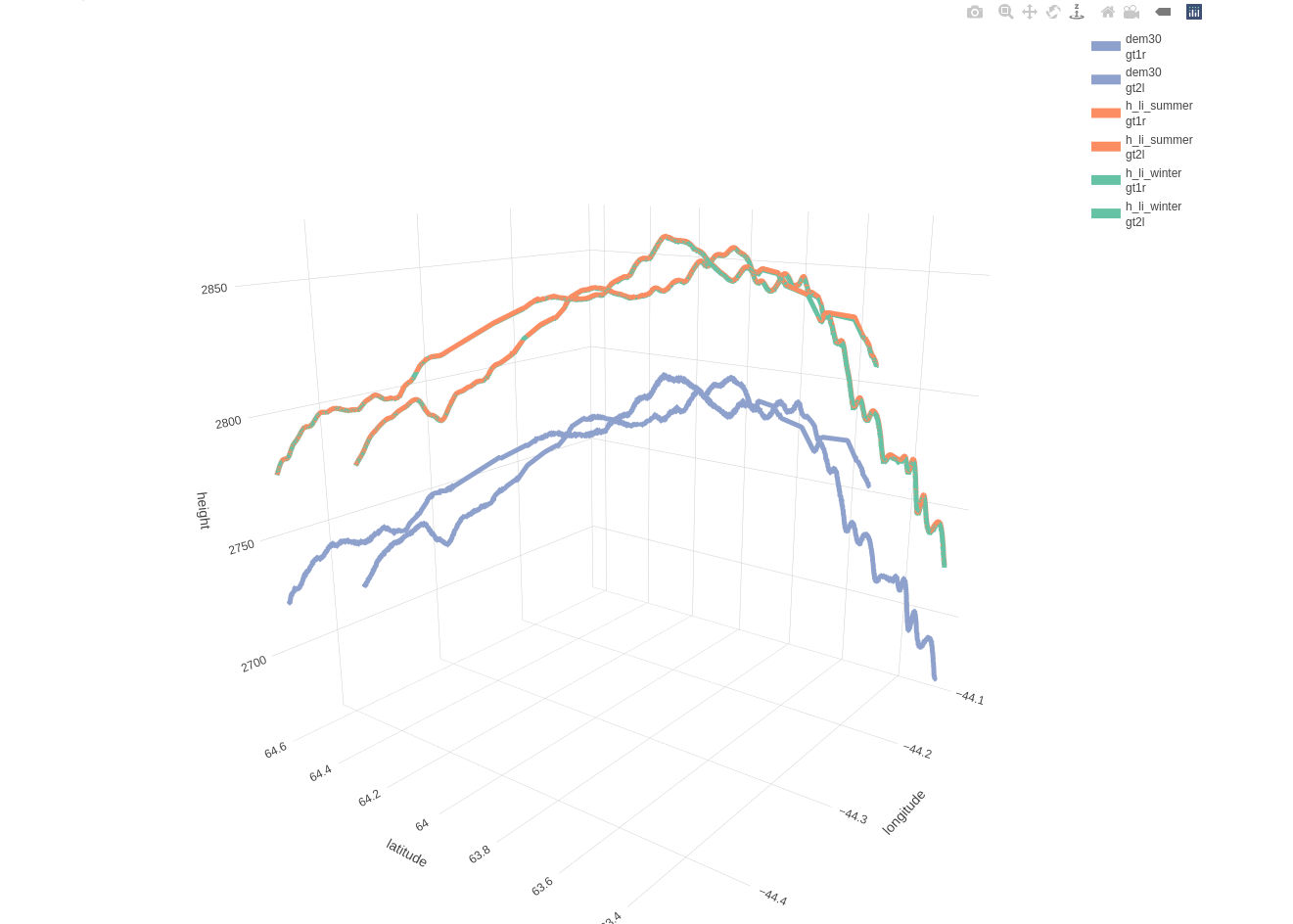
Based on the included 30-meter DEM there is a mean distance of 11.1490 and a maximum distance of 20.5394 meters between the beam and DEM coordinates. The following 3-dimensional interactive line plot shows,
- in blue color the elevation based on the DEM (compared to the 2 beams as these are separated by a 3-km distance)
- in orange color the land-ice-height measurements of the summer period (separately for ‘gt1r’ and ‘gt2l’)
- in green color the land-ice-height measurements of the winter period (separately for ‘gt1r’ and ‘gt2l’)
cols_viz_dem = c('beam', 'longitude', 'latitude', 'h_li_winter', 'h_li_summer', 'dem30')
merg_cells_viz = merg_cells[, ..cols_viz_dem]
merg_cells_viz_mlt = reshape2::melt(merg_cells_viz, id.vars = c('beam', 'longitude', 'latitude'))
merg_cells_viz_mlt = data.table::data.table(merg_cells_viz_mlt, stringsAsFactors = F)
colnames(merg_cells_viz_mlt) = c('beam', 'longitude', 'latitude', 'variable', 'height')
fig_height = plotly::plot_ly(merg_cells_viz_mlt,
x = ~longitude,
y = ~latitude,
z = ~height,
split = ~beam, # split by beam
type = 'scatter3d',
mode = 'lines',
color = ~variable,
line = list(width = 10),
width = 1000,
height = 900)
fig_height
Installation & Citation:
An updated version of the IceSat2R package can be found in my Github repository and to report bugs/issues please use the following link, https://github.com/mlampros/IceSat2R/issues.
If you use the IceSat2R R package in your paper or research please cite both IceSat2R and the original articles https://cran.r-project.org/web/packages/IceSat2R/citation.html:
@Manual{,
title = {IceSat2R: ICESat-2 Altimeter Data using R},
author = {Lampros Mouselimis},
year = {2022},
note = {R package version 1.0.0},
url = {https://CRAN.R-project.org/package=IceSat2R},
}